Yu-Lan Wang, MS;1 Kuo-Sheng Fan, MD;2 Hui-Ju Tsai, MD;3 Chin-Yu Cheng, MS;1 Chia-Kai Tan, MS;1 Ying-Yan Chen, MS;1
Hsin-Tzu Huang, MD;4 Jen-Ho Yeh, MD;5 Chuen-Sheue Chiang, PhD1,6
1 Center for Diagnostics and Vaccine Development, Centers for Disease Control, Taipei, Taiwan
2 Department of Internal Medicine, Buddhist Dalin Tzu Chi Hospital, Chiayi, Taiwan
3 Department of Internal Medicine, HePing FuYou Branch, Taipei City Hospital, Taipei, Taiwan
4 Respiratory Care Wards, Country Hospital, Taipei, Taiwan
5 Department of Internal Medicine, Taipei Jen-Chi Hospital, Taipei, Taiwan
6 School of Nursing, National Taipei University of Nursing and Health Sciences, Taipei, Taiwan
Corresponding author:
Chuen-Sheue Chiang, PhD
Center for Diagnostics and Vaccine Development
Centers for Disease Control
No. 161, Kun-Yang Street, Taipei, Taiwan
Tel.: 886-2-26531350
Fax: 886-2-27885075
cschiang10@cdc.gov.tw
ABSTRACT
Background: A probable outbreak of respiratory disease in a nursing home serving exclusively patients in a persistent vegetative state (PVS) resulted in hospitalization of eight patients.
Methods: Microbes from all PVS patients’ respiratory tracks and environments were surveyed by microbiological methods. Major pathogenic microbes were analyzed by pulsed-field gel electrophoresis (PFGE).
Results: 24 PVS patients were investigated. Half were colonized with at least four different pathogenic microbes in their respiratory tracts. The most prevalent microbes were Pseudomonas aeruginosa in 15 patients (62.5%), Serratia marcescens in 14 (58.3%), Citrobacter koseri in nine (37.5%), Streptococcus pneumoniae in six (25%), and Proteus mirabilis in five (20.8%). By PFGE analysis, one major pulsotype each was identified for S. marcescens (92.9%, 13/14) and S. pneumoniae (100%, 6/6), whereas diverse pulsotypes were identified for P. aeruginosa, C. koseri, and P. mirabilis. Both major pulsotypes for S. marcescens and S. pneumoniae were also found in strains from patients outside the nursing home. No environmental reservoir was found for prevalent microbes.
Conclusions: Clonal transmission of S. marcescens and S. pneumoniae among PVS patients in the nursing home was evident, indicating a need to enforce control measures to reduce threats to this type of facility.
KEYWORDS
Microbial surveillance; microbial transmission; persistent vegetative state; Serratia marcescens; Streptococcus pneumoniae
INTRODUCTION
Episodes of infectious disease are important issues in nursing homes, where respiratory infections are most common [1, 2]. Infections can cause high morbidity and mortality among residents [3] since conditions there are ideal for the dissemination of infectious agents. Such conditions are susceptible residents, common exposure sources, people flow, and long-term residence [4]. Development of nosocomial or healthcare-associated infections are associated with two key pathophysiological factors, colonization of pathogenic organisms and impaired host immune defense [5].
Nursing homes that serve patients in a persistent vegetative state (PVS) possess additional distinctions that predispose residents to infections. PVS patients require comprehensive daily care and hygienic practice that is fulfilled solely by nursing home staff. Most PVS patients use intruding devices, well-known risk factors associated with infectious reservoirs [5]. Surveillance studies showed that prevalence of pneumonia in nursing homes for PVS patients was 14.2% [6] and that pathogenic colonization and being susceptible to aspiration pneumonia and systemic infections were associated with tube-fed institutionalized elderly patients [7-9].
A probable outbreak of respiratory disease involving hospitalization of eight PVS patients in a nursing home alerted the health authority to prompt this study. The purpose was to survey microbe prevalence in respiratory tracts of all PVS patients to determine any person-to-person transmission of microbes. We also surveyed surrounding environments to find reservoirs for suitable control measures.
METHODS
Background and setting
Through the national surveillance system for healthcare institutions, a cluster of respiratory infection was reported to the Taiwan Centers for Disease Control in February 2009. It took place in a 45-bed nursing home in northern Taiwan serving PVS patients from low- and middle-income families by a social welfare foundation. When the outbreak occurred, a total of 25 PVS patients resided in three separate wards and received physical therapy of steam inhalation and sputum suction daily.
Microbial surveillance
Sputum was collected from 24 PVS patients. Environmental samples were taken by using swabs from all possible reservoirs, including tubes, bed railings, spraying humidifiers, faucets, shower heads, mops, water buckets, outlets of reverse osmosis (RO) water, sinks, aprons, CD carts, and telephones. Sputum samples, pretreated with sterile glass beads by vortexing, and environmental swabs were inoculated on blood, chocolate, and MacConkey agar plates. Suspected colonies were selected for identification. Bacteria species and antimicrobial susceptibility were determined by biochemical reaction agar-tubes and the Phoenix Automated Microbiology System (BD, Sparks, MD, U.S.A.) using PMIC/ID-14 and NMIC/ID-4 panels. Serotype of Streptococcus pneumoniae was determined by capsule swelling test with pneumococcal antisera (SSI, Copenhagen, Denmark). Pneumocystis carinii was detected with polymerase chain reaction [10].
Bacterial genotyping
Genotyping was performed using pulsed-field gel electrophoresis (PFGE) analysis with the CHEF-DRIII apparatus (Bio-Rad, Hercules, CA, U.S.A.). Ramp and running time were five to 50 seconds and 21 hours with SpeI digestion, or five to 15 seconds and eight hours followed by 15 to 45 seconds and 12 hours with XbaI digestion for Serratia marcescens; five to 30 seconds and 24 hours with SpeI digestion for Pseudomonas aeruginosa and Citrobacter koseri; five to 40 seconds and 23 hours with SfiI digestion for Proteus mirabilis; and two to 20 seconds and 21 hours with SmaI digestion for S. pneumoniae. BioNumerics 4.0 software (Applied Maths, Austin, TX, U.S.A.) was used to determine clonal similarity. Greater than 80% similarity in genetic relatedness was defined as strains with the same pulsotype.
Statistics
Categorical variables were analyzed using χ2 statistic or Fisher exact test. In all data analysis, a p value of < 0.05 was considered significant.
RESULTS
History and patient characteristics
The average age of all 25 PVS patients was 43.8 years (range: 15 to 87 years), with an average residence of 4.7 years (Table 1). In late January 2009, a 26-year-old male, the index case, was hospitalized due to fever, tachypnea, and pneumonia patches in lungs. Within ten days, seven more PVS patients were hospitalized due to respiratory symptoms (32.0% attack rate). They, including six males, came from all three wards, with an average age of 51.6 years (range: 23 to 67 years). Seven hospitalized PVS patients recovered within one week, and the index case had a longer hospital stay.
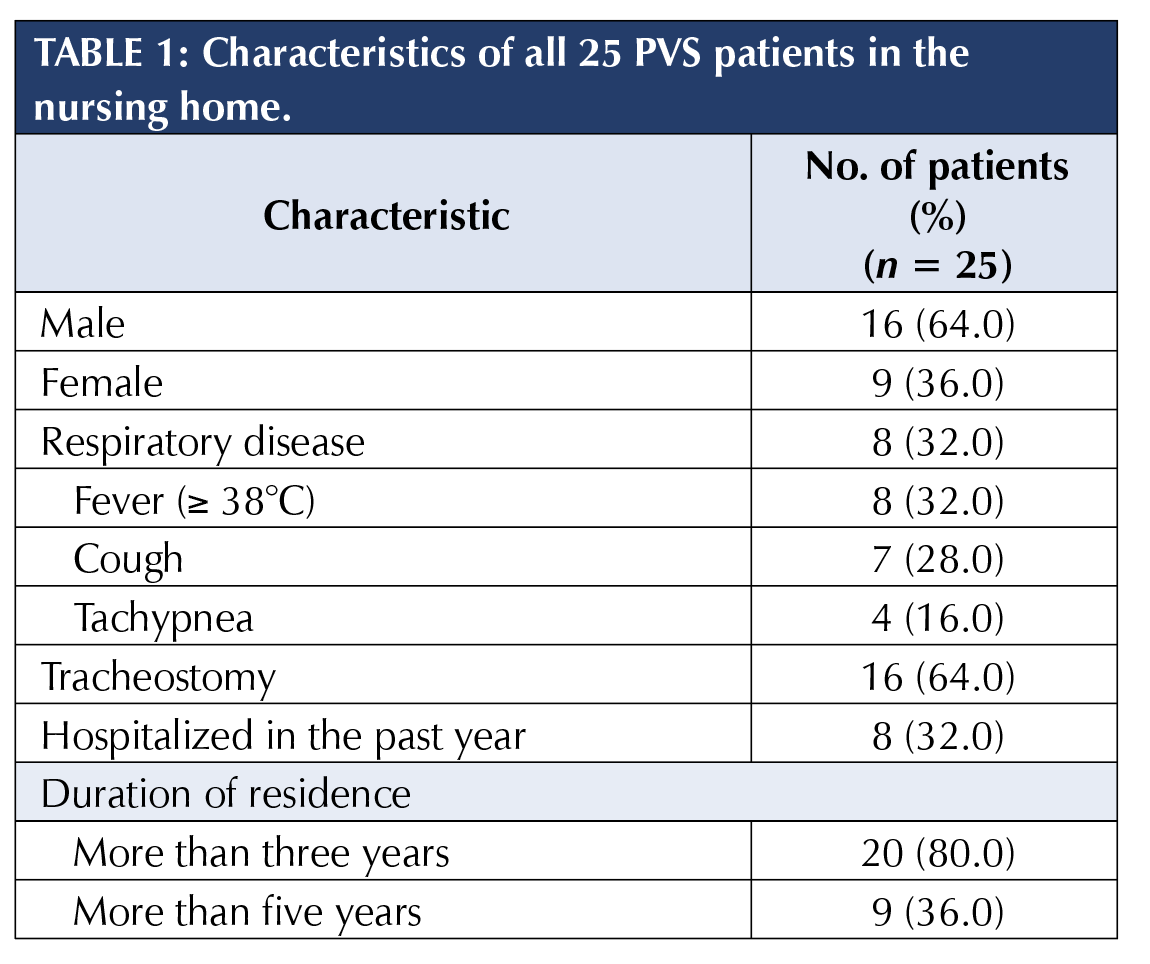
None of the 15 healthcare workers developed respiratory symptoms two weeks before and after the outbreak. Neither did visitors who visited the nursing home one week before. When performing caring duties, healthcare workers wore masks and gloves according to the standard operation protocols. All PVS patients and healthcare workers received seasonal influenza vaccine prior to the outbreak.
S. marcescens was isolated from the index case’s sputum three days after disease onset in the hospital. Nonetheless, no viral or bacterial cause was concluded. Both clinical characteristics and remedy of antibiotic treatment were not specific for S. marcescens infection. However, the outbreak was terminated in a short period due to implementation of control measures, including enhanced hand hygiene, strengthened environment cleanliness and equipment disinfection, and suspending visiting for two weeks.
Microbial surveillance
Immediately following the episode, a microbial surveillance of the respiratory tract was conducted for 24 PVS patients, excluding the index case patient, who was then still hospitalized (Table 2). As for Gram-negative bacteria (GNB), 15 PVS patients (62.5%) were colonized with P. aeruginosa; 14 patients (58.3%) were colonized with S. marcescens, including five of the seven hospitalized and recovered patients (71.4%); and nine and five patients were colonized with C. koseri and P. mirabilis, respectively. As for Gram-positive bacteria, six patients were colonized with S. pneumoniae (25%). Polymicrobial colonization was common. 50% of PVS patients were colonized with at least four different microbes.
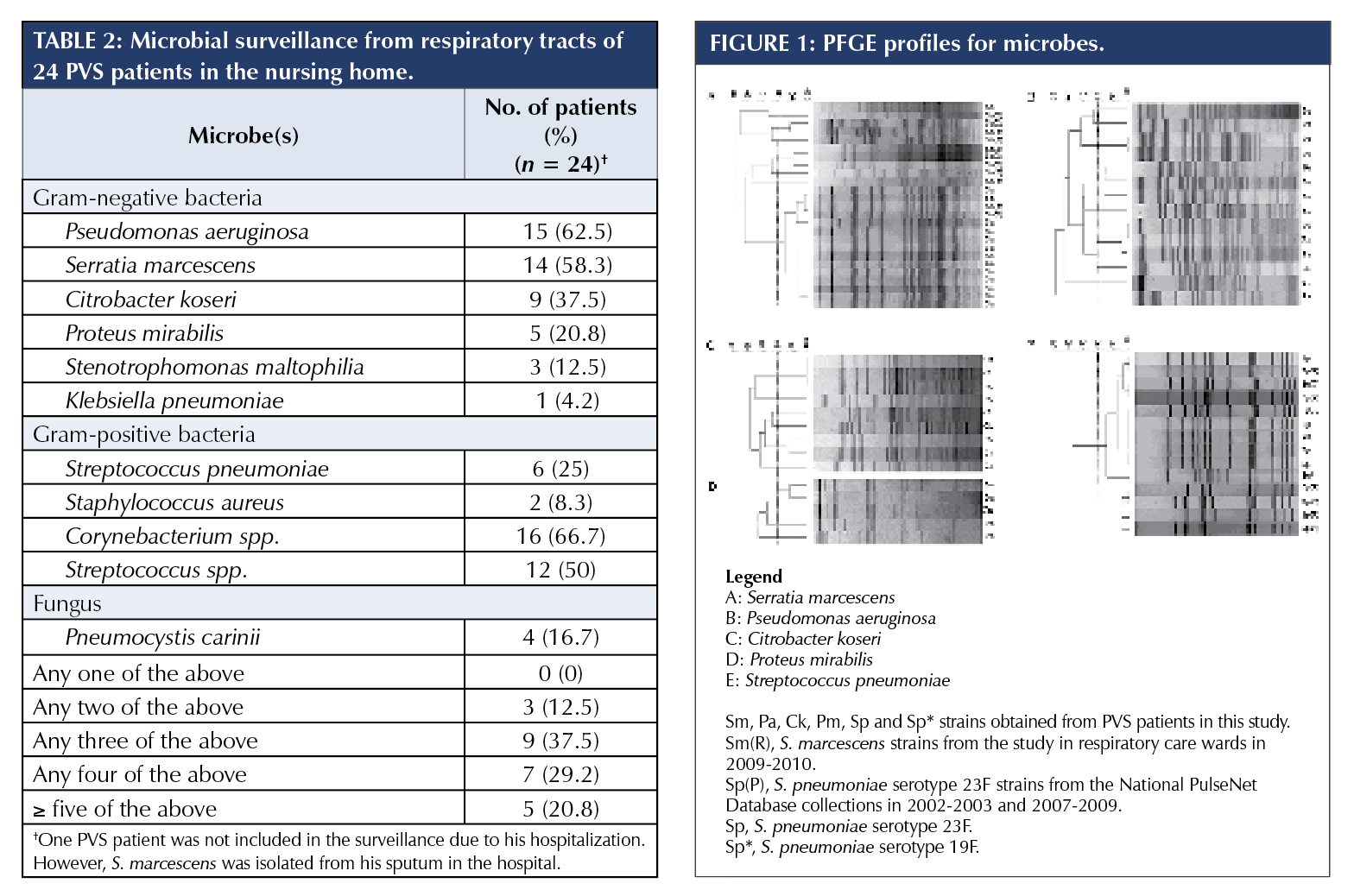
None of the variables analyzed – including age group, hospitalization in the past year, with tracheostomy, albumin level, length of residence, and P. aeruginosa colonization – was significantly associated with S. marcescens colonization.
Environmental reservoirs were not found for prevalent microbes. Only Bacillus cereus, Enterobacter cloacae, and Stenotrophomonas maltophilia were identified from the outlets of RO water, mops, and sinks in the nursing station. One of the 15 P. aeruginosa strains was resistant to imipenem (6.7%) and none of the S. marcescens strains was an extended-spectrum beta-lactamases producer. Both S. aureus strains were methicillin-resistant.
Bacterial genotyping
To clarify possible transmission in the nursing home, all 49 strains of the five major microbes were analyzed by PFGE genotyping (Figure 1). For S. marcescens, one major pulsotype was identified for 13 of the 14 strains (92.9%), including all five strains from the seven hospitalized and recovered patients (Figure 1A, SpeI digestion). Restriction digestion with XbaI gave the same result (data not shown). For P. aeruginosa, nine pulsotypes were identified for 14 of the 15 strains (Figure 1B). One P. aeruginosa strain could not be digested by SpeI. For C. koseri and P. mirabilis, six and three pulsotypes were identified, respectively (Figures 1C and 1D). For S. pneumoniae, one pulsotype was identified for all six strains (100%).
PFGE genotyping was applied to 11 S. marcescens strains collected from a microbial surveillance of patients in general respiratory care wards (RCWs) as well (see “Discussion”). Seven pulsotypes were identified, including the major pulsotype in this study, which was observed for two strains from the same hospital in southern Taiwan (Figure 1A). Meanwhile, PFGE genotypes of S. pneumoniae strains in this study were submitted to the National PulseNet Database of S. pneumoniae in Taiwan. The database included PFGE genotypes of S. pneumoniae strains isolated from patients with invasive infections throughout Taiwan during 2002-2003 and 2007-2009 (unpublished data). In the database, a total of 199 S. pneumoniae serotype 23F strains were classified into nine pulsotypes, including two major pulsotypes consisting of 90 (45.2%) and 87 (43.7%) strains, respectively. All six S. pneumoniae strains in this study shared the same pulsotype as the 90 strains from the database (Figure 1E).
DISCUSSION
In this study, we found that polymicrobial colonization was common, GNB colonization was prevalent, and person-to-person transmission of S. marcescens and S. pneumoniae was evident among PVS patients.
A microbial surveillance for patients residing in RCWs conducted in 2009-2010 revealed that 45.9% and 14.9% of patients were colonized with P. aeruginosa and S. marcescens in their respiratory tracts, respectively (our unpublished data). The S. marcescens colonization rate was significantly lower than that in PVS patients (p < 0.001), while the P. aeruginosa colonization rate was not (p = 0.16).
P. aeruginosa constituted a high proportion of pathogenic GNB from respiratory tracts of tube-fed elderly patients (31% and 34% in two studies) [7, 11]. P. aeruginosa (23.4%) and S. marcescens (10.8%) were major microbes in a bacterial surveillance for respiratory aspirates from patients in RCWs [12]. P. aeruginosa was well-known for its colonizing tendency for respiratory equipment and thriving in oropharynx. S. marcescens emerged as an opportunistic pathogen to cause outbreaks, likely attributable to its rapid spreading and innumerable heterogeneous clones, its potential reservoirs in infected or colonized carriers and inanimate objects, and its correlation with use of intruding tubes [13-19]. A previous study reported that 89% of PVS patients in Taiwan used a nasogastric tube (NGT) for feeding [6]. Most PVS patients in our study also used an NGT for feeding.
In our study, one pulsotype each was dominant for S. marcescens and S. pneumoniae. In contrast, diverse pulsotypes were identified for P. aeruginosa, C. koseri, and
P. mirabilis in our study as well as for S. marcescens from RCW patients and for S. pneumoniae from the National PulseNet Database. These results clearly suggest that S. marcescens and S. pneumoniae were transmitted among PVS patients in the nursing home. Furthermore, the dominant pulsotype for either S. marcescens or S. pneumoniae in the nursing home was not unique, since it was also found in S. marcescens strains from RCW patients and in S. pneumoniae strains from the National PulseNet Database. This result indicates that both dominant pulsotypes for the S. marcescens and S. pneumoniae strains were circulating in the community as well. Since at least 37.5% of all nosocomial infections were due to cross-transmission [20], microorganisms from outside environments constituted a great public health concern.
There were limitations to our study. First, it was carried out in one single nursing home with a small patient number. However, the entire PVS population was included, except the index case patient, and the findings represented the real situation in this facility. Second, no risk factor was found in association with S. marcescens colonization, suggesting that further studies are required. Third, the exact mode of microbial cross-transmission was not identified. The outbreak occurred during a one-week holiday; infectious reservoirs were likely eliminated during environmental disinfection. Nonetheless, clonal transmission among PVS patients was supported by bacterial genotyping results.
In conclusion, we report the cross-transmission of S. marcescens and S. pneumoniae in a nursing home serving PVS patients, highlighting the threat to this type of healthcare facility and the importance of comprehensive control measures.
REFERENCES
1. Jackson, M. M., & Fierer, J. (1985). Infections and infection risk in residents of long-term care facilities: A review of the literature, 1970-1984. American Journal of Infection Control, 13(2), 63-77.
2. Loeb, M., McGeer, A., McArthur, M., Peeling, R. W., Petric, M., & Simor, A. E. (2000). Surveillance for outbreaks of respiratory tract infections in nursing homes. Canadian Medical Association Journal, 162(8), 1133-1137.
3. Thigpen, M. C., Thomas, D. M., Gloss, D., Park, S. Y., Khan, A. J., Fogelman, V. L., Beali, B., Van Beneden, C. A., Todd, R. L., & Greene, C. M. (2007). Nursing home outbreak of invasive group A streptococcal infections caused by 2 distinct strains. Infection Control and Hospital Epidemiology, 28(1), 68-74. doi: 10.1086/508821
4. Strausbaugh, L. J., Sukumar, S. R., & Joseph, C. L. (2003). Infectious disease outbreaks in nursing homes: An unappreciated hazard for frail elderly persons. Clinical Infectious Diseases, 36(7), 870-876. doi: 10.1086/368197
5. Vincent, J. L. (2003). Nosocomial infections in adult intensive-care units. Lancet, 361(9374), 2068-2077. doi: 10.1016/S0140-6736(03)13644-6
6. Lin, L. C., Hsieh, P. C., & Wu, S. C. (2008). Prevalence and associated factors of pneumonia in patients with vegetative state in Taiwan. Journal of Clinical Nursing, 17(7), 861-868. doi: 10.1111/j.1365-2702.2006.01883.x
7. Leibovitz, A., Plotnikov, G., Habot, B., Rosenberg, M., & Segal, R. (2003). Pathogenic colonization of oral flora in frail elderly patients fed by nasogastric tube or percutaneous enterogastric tube. Journals of Gerontology: Series A, 58(1), 52-55.
8. Limeback, H. (1998). Implications of oral infections on systemic diseases in the institutionalized elderly with a special focus on pneumonia. Annals of Periodontology, 3(1), 262-275. doi: 10.1902/annals.1998.3.1.262
9. Marik, P. E. (2001). Aspiration pneumonitis and aspiration pneumonia. New England Journal of Medicine, 344(9), 665-671. doi: 10.1056/NEJM200103013440908
10. Beard, C. B., Roux, P., Nevez, G., Hauser, P. M., Kovacs, J. A., Unnasch, T. R., & Lundgren, B. (2004). Strain typing methods and molecular epidemiology of Pneumocystis pneumonia. Emerging Infectious Diseases, 10(10), 1729-1735. doi: 10.3201/eid1010.030981
11. Leibovitz, A., Dan, M., Zinger, J., Carmeli, Y., Habot, B., & Segal, R. (2003). Pseudomonas aeruginosa and the oropharyngeal ecosystem of tube-fed patients. Emerging Infectious Diseases, 9(8), 956-959. doi: 10.3201/eid0908.030054
12. Kuo, L. C., Yu, C. J., Kuo, M. L., Chen, W. N., Chang, C. K., Lin, H. I., Chen, C. C., Lu, M. C., Lin, C. H., Hsieh, W. F., Chen, L. W., Chou, Y., Huang, M. S., Lee, C. H., Chen, S. C., Thai, S. L., Chen, P. C., Chen, C. H., Tseng, C. C., Chen, Y. S., Hsiue, T. R., & Hsueh, P. R. (2008). Antimicrobial resistance of bacterial isolates from respiratory care wards in Taiwan: A horizontal surveillance study. International Journal of Antimicrobial Agents, 31(5), 420-426. doi: 10.1016/j.ijantimicag.2008.01.010
13. Fleisch, F., Zimmermann-Baer, U., Zbinden, R., Bischoff, G., Arlettaz, R., Waldvogel, K., Nadal, D., & Ruef, C. (2002). Three consecutive outbreaks of Serratia marcescens in a neonatal intensive care unit. Clinical Infectious Diseases, 34(6), 767-773. doi: 10.1086/339046
14. Casolari, C., Pecorari, M., Fabio, G., Cattani, S., Venturelli, C., Piccinini, L., Tamassia, M. G., Gennari, W., Sabbatini, A. M. T., Leporati, G., Marchegiano, P., Rumpianesi, F., & Ferrari, F. (2005). A simultaneous outbreak of Serratia marcescens and Klebsiella pneumoniae in a neonatal intensive care unit. Journal of Hospital Infection, 61(4), 312-320. doi: 10.1016/j.jhin.2005.03.005
15. Polilli, E., Parruti, G., Fazii, P., D’Antonio, D., Palmieri, D., D’Incecco, C., Mangifesta, A., Garofalo, G., Del Duca, L., D’Amario, C., Scimia, M., Cortesi, V., & Fortunato, V. (2011). Rapidly controlled outbreak of Serratia marcescens infection/colonisations in a neonatal intensive care unit, Pescara General Hospital, Pescara, Italy, April 2011. Eurosurveillance, 16(24), pii=19892. doi: https://doi.org/10.2807/ese.16.24.19892-en
16. Sikka, M. K., Hayden, M. K., Pur, S., Segreti, J., Harris, A. A., Weinstein, R. A., & Trenholme, G. (2010). Microbiologic and clinical epidemiologic characteristics of the Chicago subset of a multistate outbreak of Serratia marcescens bacteremia. Infection Control and Hospital Epidemiology, 31(11), 1191-1193. doi: 10.1086/657074
17. Chetoui, H., Delhalle, E., Melin, P., Struelens, M. J., De Ryck, R., Osterrieth, P., & De Mol, P. (1998). Typing of nosocomial strains of Serratia marcescens: Comparison of pulsed-field gel electrophoresis of macrorestriction fragments with biotyping, esterase typing and ribotyping. Research in Microbiology, 149(2), 137-143.
18. Su, L. H., Ou, J. T., Leu, H. S., Chiang, P. C., Chiu, Y. P., Chia, J. H., Kuo, A. J., Chiu, C. H., Chu, C., Wu, T. L., Sun, C. F., Riley, T. V., & Chang, B. J. (2003). Extended epidemic of nosocomial urinary tract infections caused by Serratia marcescens. Journal of Clinical Microbiology, 41(10), 4726-4732.
19. van der Sar-van der Brugge, S., Arend, S. M., Bernards, A. T., Berbee, G. A. M., Westendorp, R. G. J., Feuth, J. D. M., & van den Broek, P. J. (1999). Risk factors for acquisition of Serratia marcescens in a surgical intensive care unit. Journal of Hospital Infection, 41(4), 291-299. doi: https://doi.org/10.1053/jhin.1998.0498
20. Weist, K., Pollege, K., Schulz, I., Rüden, H., & Gastmeier, P. (2002).
How many nosocomial infections are associated with cross-transmission?
A prospective cohort study in a surgical intensive care unit. Infection Control and Hospital Epidemiology, 23(3), 127-132. doi: 10.1086/502021


