Jude Nwaokenye, MPH1, Justin M. Chan, PhD2, Xuetao Wang, MPH1, Katy Short, MPH1, Shazia Masud, MD3,5, Shannon L. Russell, PhD4,5, James Zlosnik EA, PhD4,5, Natalie Prystajecky, PhD4,5, Robert Shaw, MD6, and Calvin Cheng, MD7*
1Infection Prevention and Control Department, Fraser Health Authority, Surrey, B.C., Canada
2Department of Experimental Medicine, University of British Columbia, Vancouver, B.C., Canada
3Department of Pathology and Laboratory Medicine, Surrey Memorial Hospital, Surrey, B.C., Canada
4British Columbia Centre for Disease Control, Vancouver, B.C., Canada
5Pathology and Laboratory Medicine, University of British Columbia, Vancouver, B.C., Canada
6Department of Internal Medicine, Delta Hospital, Delta, B.C., Canada
7Department of Hospital Medicine, Delta Hospital, Delta, B.C., Canada
*Correspondence:
Calvin Cheng
5800 Mountain View Blvd,
Delta, B.C. V4K 3V6
Phone: 604 374 5772
Email: This email address is being protected from spambots. You need JavaScript enabled to view it.
ABSTRACT
Background: This study describes a SARS-CoV-2 outbreak which was declared in a community hospital, and discusses the lessons learned that could inform future outbreak prevention and control efforts.
Methods: The outbreak took place from September 16, 2020 to October 26, 2020 when COVID-19 incidence in the community was low and the COVID-19 vaccine was not yet available. Epidemiological data, patient clinical information, and whole genome sequencing were utilized for the outbreak investigation and analysis.
Results: The index case was a patient whose positive status was unknown to staff and whose symptoms on admission did not fit the screening criteria at the time. A total of 19 patients were linked to the outbreak during the study period, with an attack rate of 29%. All-cause mortality for patient cases was 37%. Whole genome sequencing confirmed genetic relatedness of all patient cases.
Conclusion: Vigilance for atypical clinical presentation, strategic patient cohorting, and minimizing movement of positive and exposed patients may help limit transmission. Whole genome sequencing can supplement epidemiological data to inform outbreak investigations.
INTRODUCTION
The global COVID-19 pandemic, caused by the highly contagious SARS-CoV-2, has resulted in more than 763 million reported cases and almost 7 million deaths worldwide as of April 19, 2023 (WHO, 2023). SARS-CoV-2 is transmitted primarily through droplets (Triggle et al., 2021) and the documented asymptomatic or pre-symptomatic transmission presents additional challenges for controlling the spread of the disease (Lee et al., 2020; Johansson et al., 2021). Healthcare transmission is especially concerning, as it often involves vulnerable patient populations at risk for severe outcomes (Gao et al., 2021; Gómez-Ochoa et al., 2020; Chen et al., 2021; Turale et al., 2021), and places significant strain on already burdened healthcare systems (Triggle et al., 2021; Kanji et al., 2022). Due to the increased vaccination coverage and the rapid spread of the Omicron variant, with less severe clinical outcomes (Ren et al., 2022), many countries have relaxed COVID-19 public health measures. However, as nosocomial transmissions and COVID-19 outbreaks continue to occur in acute care hospitals and new variants continue to emerge, learnings from outbreak investigations can help inform infection control measures.
We describe a COVID-19 outbreak that was declared on September 16, 2020 at a small suburban acute care center in British Columbia, Canada. The report aims to provide a description of the outbreak, incorporating both epidemiological and genomic data, and discusses lessons learned that could inform future outbreak prevention and control efforts.
Material and methods
Study setting and design
This cross-sectional, observational study was conducted in a 63-bed community hospital in Fraser Health (FH), the largest regional health authority in British Columbia, Canada. The outbreak involved two medical units (South Unit A, with three multi-bed rooms and two private rooms for a total of 14 beds, and North Unit, with four multi-bed rooms, one semi-private room, and one private room, for a total of 19 beds) and one alternative level of care unit (South Unit B, 10 beds across two multi-bed rooms and two private rooms). During the study period, the average community incidence was 6.16 cases per 100,000, and COVID-19 vaccines were not yet available. At the start of the pandemic, the hospital had implemented universal masking and eye protection for all healthcare workers, restricted visitation, and introduced screening for COVID-19 symptoms and exposure risk factors in the emergency department. SARS-CoV-2 polymerase chain reaction (PCR) testing using nasopharyngeal swabs was performed for all patients presenting with symptoms, a history of travel or reported exposure to COVID-19. All suspected and confirmed COVID-19-positive patients were cohorted separately on droplet and contact precautions in dedicated units with dedicated staffing and equipment. In addition, airborne precautions were applied for all aerosol-generating procedures with COVID-19 confirmed, suspected or exposed patients.
Case definitions and outbreak control measures
At the time of the outbreak, a healthcare-associated COVID-19 case was defined as laboratory-confirmed COVID-19 with the symptom onset or specimen collection date five or more days after admission. For consistency, the specimen collection date of the first positive test was used as a proxy for symptom onset. Confirmed epidemiological links were defined as any exposures without appropriate personal protective equipment (PPE), or droplet and contact precautions to a confirmed case during the period of infectivity. During the outbreak period, the infectious period was defined as two days prior to symptom onset or positive specimen collection, whichever occurred first, through to symptom resolution and a negative SARS-CoV-2 PCR test.
All SARS-CoV-2-positive specimens were reported in real-time to the hospital Infection Prevention and Control (IPC) practitioner, and a COVID-19 outbreak was declared when there was evidence of transmission involving a healthcare-associated-COVID-19 patient case on a unit (as defined by the geographical area, nursing station, and unit mnemonic). Outbreaks were declared in real-time as cases were identified. Outbreaks were declared over 14 days after the last identified exposure to a confirmed case.
Outbreak control measures included weekly SARS-CoV-2 point prevalence testing of all exposed patients, defined as unit contacts, as well as hospital-wide healthcare worker testing, enhanced cleaning and daily hand hygiene and personal protective equipment audits. Confirmed COVID-19 cases were transferred to a dedicated unit, and all unit contacts were placed on droplet and contact precautions for the duration of the outbreak. A list of all discharged unit contacts was provided to Public Health for notification and follow up in the community, and all cases identified from the discharged contacts were included in the outbreak investigation. Healthcare workers were dedicated to the outbreak unit, and the unit was closed to new admissions. Healthcare workers who tested positive for COVID-19, or were exposed to a case without appropriate PPE were excluded from work as per provincial Public Health guidance. During the outbreak period, only essential visitors were permitted, as per provincial policy. No visitor cases of COVID-19 were identified during the outbreak investigation.
Comprehensive case assessments and contact tracing was completed for all COVID-19 cases identified within the FH region by regional IPC and Public Health. All cases were reviewed by both Public Health and IPC for association with the hospital outbreak. For this study, a retrospective patient chart review was conducted through the hospital electronic medical record system.
Whole genome sequencing
All COVID-19 specimens associated with the outbreak were sent to the British Columbia Centre for Disease Control Public Health Laboratory for whole genome sequencing (WGS). Detailed WGS laboratory methods have been described previously (Hickman et al., 2020). Briefly, samples were sequenced on an Illumina MiSeq instrument and analyzed using a modified ARCTIC Nextflow pipeline (Hickman et al., 2020). Sequences passing quality control were included in the phylogenetic analysis. Phylogenetic trees were constructed using Nextstrain (Hadfield et al., 2018), and samples were manually assigned to a genetic clade based on an inclusion criterion of three mutations or less. Sequences differing by zero mutations were considered “Identical”, one to two mutations “Nearly Identical”, three mutations “Similar” and greater than three mutations “Different”. Samples were assigned a sub-clade designation (e.g., Clade 1.1) to denote clusters of genetically identical sequences. Lineage assignment was performed using the Phylogenetic Assignment of Named Global Outbreak Lineages tool (Pangolin Version V.3.1.17) (Rambaut et al., 2020). Results from the WGS analysis were then linked to epidemiological information.
The study was approved by the Fraser Health Research Ethics Board.
Results
Transmission from index case to other patients on the unit and healthcare workers
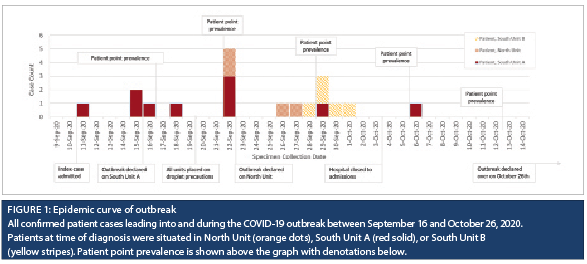
The index case (P1) resided at a community independent living facility. P1 first presented to the emergency department on September 11, 2020, with complaints of generalized body pain and a recent fall. A few hours after presentation, she was admitted into a shared room in South Unit A. P1 had no respiratory symptoms, but did have some history of loose stools at the community facility. A few hours after admission into the shared room, P1 was noted to have active diarrhea. Shortly after, healthcare workers were notified that the patient had been tested for SARS-CoV-2 prior to admission at the community facility and the test had resulted positive. The patient was immediately transferred to a private room and placed on droplet and contact precautions. Two roommates (P2 and P3) were identified as exposed, placed on similar precautions as the index patient, and the remaining empty beds in the room were closed. The index case, P1, had spent nine hours in the multi-patient room and would have used the washroom with P3; however, P2 was not mobile and had a dedicated commode. Two days after exposure to P1, P2 and P3 developed symptoms and were tested for SARS-CoV-2. Both resulted positive, and a COVID-19 outbreak was declared for South Unit A on September 16, 2020. Thirteen patients in the impacted unit were placed on droplet and contact precautions and tested for SARS-CoV-2. The timeline of these events can be seen in Figure 1.
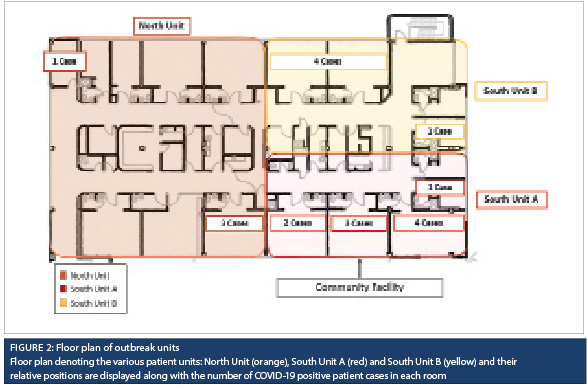
The first round of point prevalence testing identified one positive patient, P4. Two days following, another patient (P5) became symptomatic and tested positive. Both P4 and P5 were admitted in South Unit A (Figure 2). Due to the overlap of healthcare workers between different units, enhanced cleaning and additional hand hygiene audits were implemented for South Unit B and North Unit, and a second round of patient point prevalence testing was conducted, which included patients on all three units. Two new positive patients were identified on the North Unit, and the outbreak declaration extended to South Unit B and the North Unit.
Subsequent weekly patient point prevalence testing as well as testing of symptomatic patients identified additional cases. Over a period of 40 days, 19 patients tested positive for SARS-CoV-2. Weekly hospital-wide healthcare worker point prevalence testing, as well as testing of symptomatic healthcare workers, identified 29 COVID-19-positive staff, all of whom had identified epidemiological links with outbreak-associated patient or healthcare worker cases during their incubation period, defined as a 14-day period to symptom onset or positive specimen collection. The outbreak was declared over on October 26.
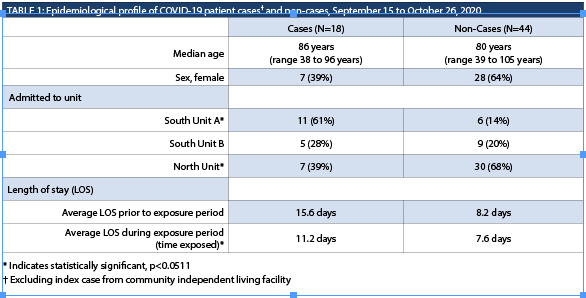
Patient demographics and clinical outcomes
A total of 62 patients were admitted to the units during the outbreak, 18 of whom tested positive for COVID-19 (excluding the index case), for an overall attack rate of 29% (95% CI: 19-41%). While the median age for cases (excluding the index case) was higher than for non-cases, and males were at a slightly higher risk than females, the differences were not statistically significant (Table 1). Patients who stayed on South Unit A were at a higher risk of contracting COVID-19, while patients who stayed on North Unit were at a lower risk. Cases had a higher average number of days at risk on an outbreak unit prior to specimen collection (or discharge) than non-cases.
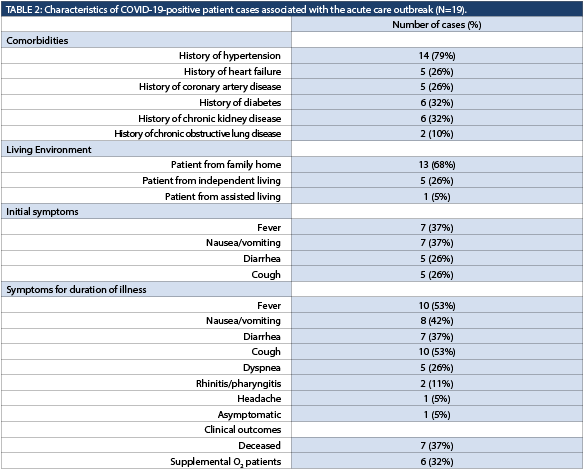
On the advance care directives of the 19 positive patients, nine had opted for all appropriate critical care interventions (full code or CPR-C2), seven wanted only medical treatment in an acute care facility (DNR-M3), two had chosen comfort care (DNR M1/M2), and one did not have a documented code status. Two of the full code patients were changed to DNR-M3 code status, one of whom later died. Thirteen of the COVID-19-positive patients lived in a family home, five resided at an independent living facility, and one from an assisted living facility. The most common comorbidities were hypertension, coronary artery disease, congestive heart failure, chronic kidney disease, and diabetes (Table 2), consistent with age demographics.
Seven out of the 19 patient cases had died by day 30 after testing positive, for an all-cause case mortality rate of 37%. Of the patients who survived, the average total length of stay was 48 days, with a range of seven to 102 days. Of the 13 patients who resided in a family home before admission, four died, one was transferred to a higher level of care, one was transferred to high-intensity rehabilitation, four were transferred to long-term care, and three were discharged.
The most common first-presenting symptoms were fever and nausea or vomiting. Six of the patients required oxygen to keep their oxygen saturation above 90%, with the maximum oxygen being 7 LPM via nasal prongs. Of the seven patients who died, only two required oxygen before their deaths. All patients who required oxygen were placed on what was standard COVID-19 treatment at the time with 6 mg dexamethasone for 10 days.
Whole genome sequencing
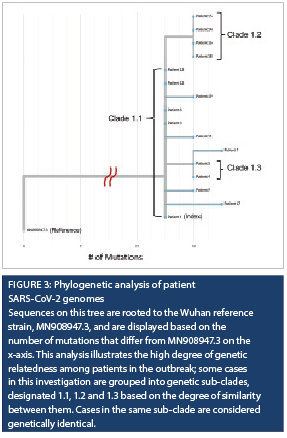
Sixteen of the 19 patient cases linked to the outbreak were successfully sequenced and were all genetically related by two mutations or less from the index case (Figure 3). Twenty-five of the 29 healthcare worker cases were also successfully sequenced, and 24 were genetically related to the index case. All cases belonged to lineage B.1, which was the most common SARS-CoV-2 lineage circulating in the province at the time.
Discussion
Over a period of 40 days, 29% of the exposed patients acquired COVID-19 in three adjacent units. Patient movements for the purpose of cohorting may have contributed to the spread of SARS-CoV-2 to adjacent units by relocating pre-symptomatic patients who tested negative on a point prevalence testing to rooms with susceptible patients. Therefore, movement of patients in facilities with limited private rooms should be carefully considered to reduce the risk of potential exposure. Additionally, the layout of the units was not conducive to cohorting and it was challenging to achieve complete physical separation of the three units during the outbreak. Although the unit, in consultation with IPC, achieved functional separation of the different cohorts, infrastructural challenges, such as shared hallways, limited private rooms, and shared supply rooms, cannot be completely omitted.
Point prevalence testing of exposed patients and healthcare workers during a COVID-19 outbreak assisted in identifying pre-symptomatic and asymptomatic cases. Asymptomatic cases have been described in literature as a veritable source of transmission, so identification of such cases allows for implementation of effective IPC measures to aid containment (Abdelmoniem et al., 2021). However, point prevalence testing can only identify cases at the time of testing, and cannot differentiate those who are truly negative and those who have acquired SARS-CoV-2, but were still incubating. Also, the results for the point prevalence should be interpreted in light of other factors such as the wider community incidence and prevalence. At the time of the outbreak, community prevalence was relatively low and as such, each case identified was more likely to represent a true infection as opposed to residual viral material from a previous community acquired SARS-CoV-2 infection.
The index case presented with diarrhea; subsequently, diarrhea was observed as an initial symptom in 26% of patients. At the time of the outbreak, nausea, vomiting and diarrhea, in the absence of respiratory symptoms, did not meet the criteria for a screening test for COVID-19. The criteria have since been updated, but this atypical presentation may have contributed to the initial rapid spread of infection as reported in literature by others (Amico et al., 2020). The poor outcomes in many of the patients were surprising as the majority did not require supplemental oxygen. This context underscores the impact of the disease at a time when less severe variants such as Omicron were not yet widespread, and vaccines were not yet available.
The patient case 30-day, all-cause mortality rate of 37% (7/19) was high, but was comparable with data from a few previous acute care outbreaks with similar patient demographics. For example, the patient case mortality rate was 35% in a hospital-wide outbreak in Edmonton, Canada which involved 31 patient cases with a mean age of 79 years (Kanji et al.., 2022), and 57% in a large hospital in Portugal which involved 21 patient cases with a median age of 82 years (Borges et al., 2021). On the other hand, a systematic review of COVID-19 case fatality rates based on all relevant studies published in 2020 gave a lower-case fatality rate of 13% among hospitalized patients and 37% among patients in the ICU (Alimohamadi et al., 2021). The studies included in the systematic review had lower median/mean age among COVID-19 patient cases, ranging from 37 to 72 years. Therefore, advanced age and associated comorbidities likely contributed to the high mortality rate among cases in these acute care outbreaks (Tahvildari et al., 2021).
Whole genome sequencing provided valuable information to the investigation and validated the epidemiological data, linking all subsequent patient cases and all but one of the healthcare worker cases to the admission of the index case. Cases genetically related to the index patient signified a very high likelihood that these infections were from a common source at a time when community prevalence was low. In settings where community prevalence is high, whole genome sequencing alone may not provide adequate evidence for a common source, since multiple introductions of the same strain is possible. It is, therefore, important to link genomic data with epidemiological information for proper interpretation.
Strengths and limitations
The case report provides unique insight into an outbreak in a community hospital, which is a setting not often represented in current literature. Whole genome sequencing, in addition to the epidemiological data, provided supporting evidence to the nosocomial spread of COVID-19 in this acute care setting. The outbreak occurred at a time when COVID-19 community prevalence was low, vaccination was not available, and before the emergence of COVID-19 variants of concern, so may not be representative of outbreaks from other variants. However, the learnings from this outbreak, including vigilance on atypical presentation and the effectiveness of outbreak interventions, can be applied to all COVID-19 outbreaks.
While 29 healthcare workers tested positive during this outbreak, the focus of this case report was limited to patient cases due to data availability and possible exposures outside of the workplace for staff. During the course of the outbreak, healthcare worker PPE use, social distancing practices, and hand hygiene compliance were explored as part of the investigation, however, limited observational data is available. Understanding risk factors for COVID-19 infection among healthcare workers is critical to informing prevention measures and additional research is needed to contribute to the available evidence (Dzinamarira et al., 2022).
Published literature indicates that Heating, Ventilation and Air Conditioning (HVAC) may play a role in the spread of SARS-CoV-2 (Thornton et al., 2022). Air quality and ventilation were considered as part of the outbreak investigation, however, there was insufficient evidence to form any conclusions. Further studies in this area would be beneficial.
CONCLUSION
Despite due diligence among healthcare workers and rapid implementation of infection control measures, epidemiological and genomic data indicate rapid nosocomial spread of COVID-19 among patients over a 40-day period across three hospital units. As the epidemiology of SARS-CoV-2 continues to evolve, we acknowledge the challenges experienced by acute care facilities and hope this may serve as a reference for how quickly an outbreak of an emerging pathogen can develop.
REFERENCES
1. World Health Organization. WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ [Accessed 22nd August 2022].
2. Triggle, C.R., Bansal, D., Ding, H., Islam, M.M., Farag, EABA, Hadi, H,A,, et al. (2021). A Comprehensive Review of Viral Characteristics, Transmission, Pathophysiology, Immune Response, and Management of SARS-CoV-2 and COVID-19 as a Basis for Controlling the Pandemic. Frontiers in Immunology. https://doi.org/10.3389/fimmu.2021.631139.
3. Lee, S., Meyler, P., Mozel, M., Tauh, T., Merchant, R. (2020). Asymptomatic carriage and transmission of SARS-CoV-2: What do we know?. Canadian Journal of Anesthesia. p. 1424–1430. https://doi.org/10.1007/s12630-020-01729-x.
4. Johansson, M.A., Quandelacy, T.M., Kada, S., Prasad, P.V., Steele, M., Brooks, J.T., et al. (2021). SARS-CoV-2 Transmission from People without COVID-19 Symptoms. Journal of the American Medical Association Network Open, 4(1). https://doi.org/10.1001/jamanetworkopen.2020.35057.
5. Gao, Y. D., Ding, M., Dong, X., Zhang, J. jin, Kursat Azkur, A., Azkur, D., et al. (2021). Risk factors for severe and critically ill COVID-19 patients: A review. Allergy: European Journal of Allergy and Clinical Immunology, 76(2), 428–455.
https://doi.org/10.1111/all.14657.
6. Gómez-Ochoa, S., Franco, O.H., Rojas, L., Raguindin, P.F., Roa-Diaz, Z.M., Minder, B., et al. (2020). COVID-19 in Healthcare Workers: A Living Systematic Review and Meta-analysis of Prevalence, Risk Factors, Clinical Characteristics, and Outcomes. American Journal of Epidemiology, 190(1): 161-175. 2020; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7499478.
7. Chen, Y., Klein, S.L., Garibaldi, B.T., Li, H., Wu, C., Osevala, N.M., et al. (2021). Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Research Reviews, 65: 101205.
8. Turale, S., Nantsupawat, A. (2021). Clinician mental health, nursing shortages and the COVID-19 pandemic: Crises within crises. International Nursing Review, 68(1): 12-14. https://doi.org/10.1111/inr.12674.
9. Kanji, J.N., Chan, Y.L.E., Boychuk, L.R., Boyington, C., Turay, S., Kobelsky, M., et al. (2022). SARS-CoV-2 outbreak in a Canadian suburban tertiary hospital necessitating full facility closure: a descriptive observational study. Canadian Medical Association Journal Open, 10(1), E137–E145. https://doi.org/10.9778/cmajo.20210064.
10. Ren, S.Y., Wang, W.B., Gao, R.D., Zhou, A.M. (2022). Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance.World Journal of Clinical Cases, 10(1), 1–11.
https://doi.org/10.12998/wjcc.v10.i1.1.
11. Hickman, R., Nguyen, J., Lee, T.D., Tyson, John, R;.,Azana, R., Tsang, F., Hoang, L., Prystajecky N. (2020). Rapid, High-Throughput, Cost Effective Whole Genome Sequencing of SARS-CoV-2 Using a Condensed One
Hour Library Preparation of the Illumina DNA Prep Kit. medRxiv, (165):1–13.
12. Hadfield, J., Megill, C., Bell, S.M., Huddleston, J., Potter, B., Callender, C., et al. (2018). NextStrain: Real-time tracking of pathogen evolution. Bioinformatics, 34(23): 4121–4123. https://doi.org/10.1093/bioinformatics/bty407.
13. Rambaut, A., Holmes, E.C., Toole, Á.O., Hill, V., Mccrone, J.T., Ruis, C., et al. (2020). A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiology, 5(11): 1403–1407.
https://doi.org/10.1038/s41564-020-0770-5.A.
14. Abdelmoniem, R., Fouad, R., Shawky, S., Amer, K., Elnagdy, T.(2021). SARS-CoV-2 infection among asymptomatic healthcare workers of the emergency department in a tertiary care facility. Journal of Clinical Virology, 134, 104710.
15. Amico, F.D., Baumgart, D.C., Danese, S., Peyrin-biroulet, L. (2020). Diarrhea During COVID-19 Infection : Pathogenesis. Clinical Gastroenterology and Hepatology, 1–10. https://doi.org/10.1016/j.cgh.2020.04.001.
16. Borges, V., Isidro, J., Macedo, F., et al. (2021). Nosocomial Outbreak of SARS-CoV-2 in a “Non-COVID-19” Hospital Ward: Virus Genome Sequencing as a Key Tool to Understand Cryptic Transmission. Viruses, 13: 604.
17. Alimohamadi, Y., Tola, H.H., Abbasi-Ghahramanloo. A., Janani. M., Sepandi. M. (2021). Case fatality rate of COVID-19: A systematic review and meta-analysis. Journal of Preventive Medicine and Hygiene, 62(2): E311–E320.
https://doi.org/10.15167/2421-4248/jpmh2021.62.2.1627.
18. Tahvildari, A., Arbabi, M., Farsi. Y., Jamshidi, P., Hasanzadeh, S., Calcagno, T.M., et al. (2020). Clinical features, diagnosis, and treatment of COVID-19 in hospitalized patients: A systematic review of case reports and
case series. Frontiers in Medicine, 7, 1–10. https://doi.org/10.3389/fmed.2020.00231.
19. Dzinamarira, T., Nkambule, S.J., Hlongwa, M., Mhango, M., Iradukunda, P.G., Chitungo, I., et al. (2022). Risk Factors for COVID-19 Infection Among Healthcare Workers. A First Report From a Living Systematic Review and Meta-Analysis. Safety and Health at Work, 13(3), 263–268. https://doi.org/10.1016/j.shaw.2022.04.001.
20. Thornton, G.M., Fleck, B.A., Dandnayak, D., Kroeker, E., Zhong, L., Hartling, L. (2022). The impact of heating, ventilation and air conditioning (HVAC) design features on the transmission of viruses, including the 2019 novel coronavirus (COVID-19): A systematic review of humidity. PLoS One, 17(10), 1–23. https://doi.org/10.1371/journal.pone.0275654.


